Tissue Patterning and Morphogenesis
 The main interest of our laboratory is to understand mechanisms underlying cell fate determination by cell signaling. We focus on the two highly conserved signaling pathways, the bone morphogenetic protein (BMP) and epidermal growth factor receptor (EGFR), and use fruit fly egg formation, oogenesis, as a model system.
The main interest of our laboratory is to understand mechanisms underlying cell fate determination by cell signaling. We focus on the two highly conserved signaling pathways, the bone morphogenetic protein (BMP) and epidermal growth factor receptor (EGFR), and use fruit fly egg formation, oogenesis, as a model system.
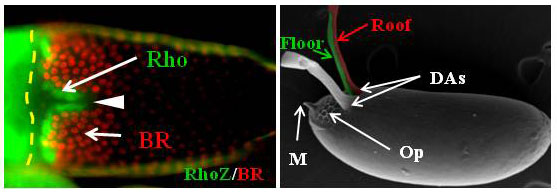
The egg chamber (left) is the precursor of the mature egg (right). The oocyte is surrounded by a mono-layer of follicle cells (FCs), which forms the 3D intricate structures of the Drosophila eggshell. Eggshell structures, such as the dorsal appendages (DAs), are formed by two non-overlapping populations of cells. Cells expressing Rhomboid (Rho) form the future floor of the DAs, and cells expressing Broad (BR) form the future roof of the DAs. Other structures include the operculum (Op – an opening for larvae hatching), and a micropyle (M – the point of sperm entry).
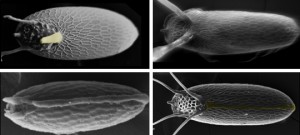 Eggshell structures are highly diverse among fly species. Morphological diversity includes the numbers and shapes of the DAs, and the lumen-like dorsal ridge and respiratory stripe.
Eggshell structures are highly diverse among fly species. Morphological diversity includes the numbers and shapes of the DAs, and the lumen-like dorsal ridge and respiratory stripe.
We are interested to understand the evolution of BMP signaling dynamics across Drosophila species, that is regulated by the pattern of the type I BMP receptor spatial expression. In addition, we aim to understand how spatiotemporal changes in EGFR activation are regulated by different distributions of Gurken, a TGF-alpha-like ligand.
Funding


