The regulation of tandem paralogs
The mammalian homologs of the Tbx-20 in Drosophila, Midline and H15, are regulated by the ETS transcription factor Pointed, which is regulated by EGFR and Jak/STAT signaling pathways. Midline and H15 are a product of gene duplication; the two the two genes reside about 55Kb away on the same chromosome. We found that the two genes are regulated by a shared enhancer, reminiscent of the bacterial operon. Specifically, an enhancer positioned at the 5′ proximal of Midline only partially regulates Midline, however, its removal completely abolishes H15 expression in the posterior end of the egg chamber.
Quantitative analysis of EGFR in epithelial tissues
To bridge the gap between qualitative and quantitative analyses of the epidermal growth factor receptor (EGFR) in tissues, we generated  an sfGFP-tagged EGF receptor (EGFR-sfGFP) in Drosophila. The homozygous fly appears similar to wild type with EGFR expression and activation patterns that are consistent with previous reports in the ovary, early embryo, and imaginal discs. Using ELISA, we quantified an average of 1100, 6200 and 2500 receptors per follicle cell (FC) at stages 8/9, 10 and ≥11 of oogenesis, respectively. Interestingly, the spatial localization of the EGFR to the apical side of the FCs at early stages depended on the TGFα-like ligand Gurken. At later stages, EGFR localized to basolateral positions of the FCs. Finally, we followed the endosomal localization of EGFR in the FCs. The EGFR colocalized with the late endosome, but no significant colocalization of the receptor was found with the early endosome. The EGFR-sfGFP fly is an exciting new resource for studying cellular localization and regulation of EGFR in tissues.
an sfGFP-tagged EGF receptor (EGFR-sfGFP) in Drosophila. The homozygous fly appears similar to wild type with EGFR expression and activation patterns that are consistent with previous reports in the ovary, early embryo, and imaginal discs. Using ELISA, we quantified an average of 1100, 6200 and 2500 receptors per follicle cell (FC) at stages 8/9, 10 and ≥11 of oogenesis, respectively. Interestingly, the spatial localization of the EGFR to the apical side of the FCs at early stages depended on the TGFα-like ligand Gurken. At later stages, EGFR localized to basolateral positions of the FCs. Finally, we followed the endosomal localization of EGFR in the FCs. The EGFR colocalized with the late endosome, but no significant colocalization of the receptor was found with the early endosome. The EGFR-sfGFP fly is an exciting new resource for studying cellular localization and regulation of EGFR in tissues.
The evolution of BMP signaling
The bone morphogenetic protein (BMP) signaling is a highly conserved pathway from worm to human. 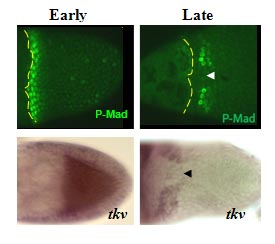 The BMP signaling (monitored by P-MAD) is dynamic in the follicle cells during oogenesis. Early signaling is along the anterior-posterior (AP) axis, and late signaling acquires a clear dorsal-ventral (DV) polarity, shown as two domains symmetrically distributed on both sides of the dorsal midline (Green, arrow head marks the midline).
The BMP signaling (monitored by P-MAD) is dynamic in the follicle cells during oogenesis. Early signaling is along the anterior-posterior (AP) axis, and late signaling acquires a clear dorsal-ventral (DV) polarity, shown as two domains symmetrically distributed on both sides of the dorsal midline (Green, arrow head marks the midline).
The dynamics of BMP signaling reflect the changes in the BMP type I receptor Thickveins (Tkv). The early uniformly expressed tkv appears later as two dorso-lateral patches on both sides of the midline (tkv in situ hybridization). TKV is necessary for BMP signaling and eggshell morphogenesis.
Dorsal structures on the Drosophila eggshell are highly diverse among species. The most prominent structures are the tube-like dorsal appendages that are different in numbers and shapes among species (A”, B”, C”). For example, D. melanogaster has two (A”), D. busckii has four (B”) and D. guttifera has three DAs (C”).
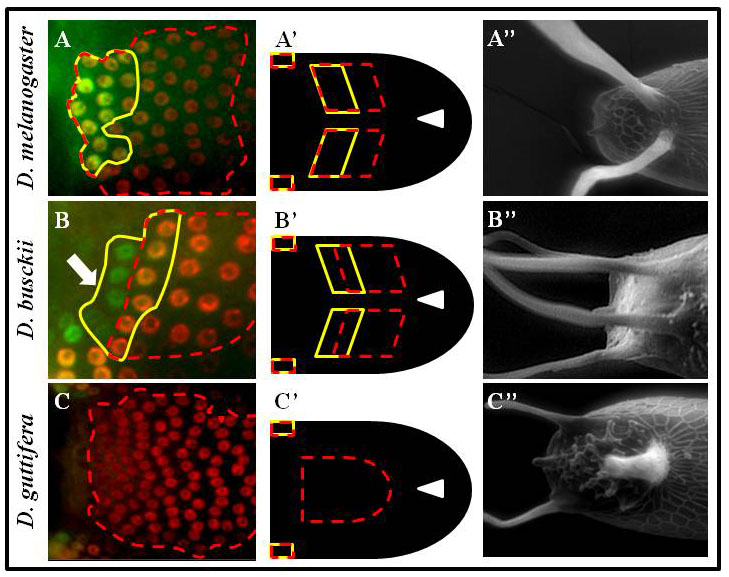 We hypothesized that this diversity reflects changes in the pattern of BMP signaling. Indeed, in reference to the position of the future roof domain of the appendage (marked by BR in red, and broken red line), the pattern of BMP signaling (green and marked by the yellow line) is different among species. The BMP signaling overlaps the BR domain in D. melanogaster (A, A’), or in addition to overlapping BR it appears in the adjacent domain in D. busckii (B, B’), or be absent from the BR domain in D. guttifera (C, C’). Remarkably, we found that differences in BMP signaling among species are determined by the pattern of tkv.
We hypothesized that this diversity reflects changes in the pattern of BMP signaling. Indeed, in reference to the position of the future roof domain of the appendage (marked by BR in red, and broken red line), the pattern of BMP signaling (green and marked by the yellow line) is different among species. The BMP signaling overlaps the BR domain in D. melanogaster (A, A’), or in addition to overlapping BR it appears in the adjacent domain in D. busckii (B, B’), or be absent from the BR domain in D. guttifera (C, C’). Remarkably, we found that differences in BMP signaling among species are determined by the pattern of tkv.
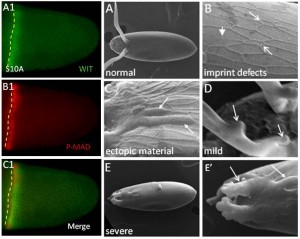 While TKV is necessary for BMP signaling, it is not sufficient. We found the type II BMP receptor, Wishful thinking (WIT), is expressed in an early P-MAD overlapping pattern (A1, B1, C1). WIT is necessary for BMP signaling, follicle cells’ patterning, eggshell morphogenesis and development (A-E’).
While TKV is necessary for BMP signaling, it is not sufficient. We found the type II BMP receptor, Wishful thinking (WIT), is expressed in an early P-MAD overlapping pattern (A1, B1, C1). WIT is necessary for BMP signaling, follicle cells’ patterning, eggshell morphogenesis and development (A-E’).
Additional genes have been associated with shaping BMP signaling in tissues. Currently, we study other components of the pathway to determine their involvement in the dynamics, intensities, and shapes of the BMP signaling in epithelial tissues.
Analysis of tissue patterning
Changes in gene regulation are associated with the evolution of morphologies. However, the specific sequence information controlling gene expression is largely unknown and discovery is time and labor consuming. We used the intricate patterning of follicle cells to probe species’ relatedness, in absence of sequence information. We focused on one of the major families of genes that pattern the Drosophila eggshell, the Chorion protein (Cp). We systematically screened for the spatiotemporal patterning of all nine Cp genes in three species: D. melanogaster, D. nebulosa, and D. willistoni (A-C). 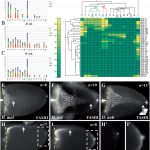
Applying an annotation code, we transformed the data into binary matrices (D) that captures the complexity of gene expression. Gene patterning is sufficient to predict species’ relatedness, consistent with their phylogeny (E). Surprisingly, we found that expression domains of most genes are different among species, suggesting that Cp gene regulation is rapidly evolving.
In addition, we found a morphological novelty along the dorsal most side of the eggshell, the dorsal ridge (DR, in B and C). Our matrix analysis placed the dorsal ridge domain in a cluster of epidermal growth factor receptor (EGFR) associated domains. We are currently studying the mechanism underlying dorsal ridge formation. 
In a recent screen for cis-regulatory modules, we discovered new domains for gene expression. Some of these patterns recapitulated the partially or fully the endogenous associated gene-pattern. These lines are a new resource for GAL4 drivers to manipulate genes during oogenesis.